Where does strain energy come
from?
(According to Molecular Mechanics)
The following table comes from Molecular Mechanics calculations
on different conformations of butane (executed using the computer program Chem3DPro). The conformations are
energy-minimized anti and gauche (note that the C-C-C-C angle in
minimized gauche is not 60°), and the energy maxima (which we'll
soon be calling passes, or transition structures, or transition
states) between them.
In the second set of entries ("Relative to Anti") the values for the
lowest energy (anti) conformation is subtracted to show how the
others differ from it. It is worth looking at how the strain energy
is distributed. There is always a give and take among different types
of distortion - one can reduce van der Waals repulsion by bending or
stretching bonds, for example. At first it seems curious that the
order of strain energy
(highlighted for
gauche) is commonly torsion > bending >
stretching, since the force constants are such that the amount of
strain energy for a given amount of atomic motion is in the opposite
order (easier to twist than to bend than to stretch). This makes sense when you think twice, because the stiffness
of the higher force constants suppresses motion and most energy
get stored in "softer" modes of distortion. [If this seems
unnatural to you click
here for an explanation.]
Below the two summary tables are the structure of
the fully eclipsed conformation and a table of its individual strain
contributions.
|
Name
|
anti
|
gauche
|
eclipsed
|
fully eclipsed
|
|
|
|
|
(anticlinal)
|
(synclinal)
|
|
C-C-C-C Angle
|
180°
|
64.04°
|
119.8°
|
0°
|
|
Stretch
|
0.1542
|
0.1725
|
0.1956
|
0.2301
|
|
Bend
|
0.2977
|
0.6064
|
0.4744
|
1.4262
|
|
Stretch-Bend
|
0.0543
|
0.0741
|
0.078
|
0.1258
|
|
Torsion
|
0.0073
|
0.4323
|
2.3682
|
2.3168
|
|
Non-1,4 VDW
|
-0.4086
|
-0.3496
|
-0.3151
|
-0.0523
|
|
1,4 VDW
|
2.0688
|
2.1022
|
2.7256
|
2.8492
|
|
Total
|
2.1737
|
3.0379
|
5.5266
|
6.8957
|
|
|
|
|
|
|
|
Relative to Anti
|
|
|
|
|
|
Stretch
|
0
|
0.0183
|
0.0414
|
0.0759
|
|
Bend
|
0
|
0.3087
|
0.1767
|
1.1285
|
|
Stretch-Bend
|
0
|
0.0198
|
0.0237
|
0.0715
|
|
Torsion
|
0
|
0.425
|
2.3609
|
2.3095
|
|
Non-1,4 VDW
|
0
|
0.059
|
0.0935
|
0.3563
|
|
1,4 VDW
|
0
|
0.0334
|
0.6568
|
0.7804
|
|
Total
|
0
|
0.8642
|
3.3529
|
4.722
|
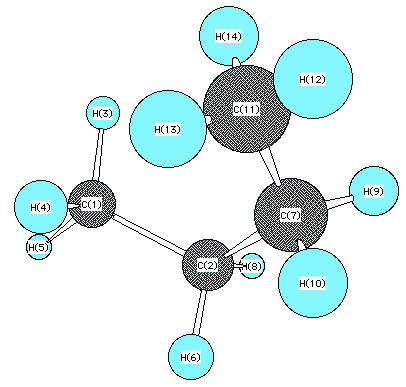
Here is a tabulation steric energy components for the
fully eclipsed conformation shown (very slightly different from the
model used for the composite table above) followed by the individual
contributions to each components. Columns headed with K(*)
are force constants for the * distortion. Within each component the individual energies are sorted to
be in decreasing order, so it is useful to look at the first few
(which are
Highlighted)
to see which atoms they are coming from.
|
Stretch
|
0.2292
|
|
|
|
|
|
|
Bend
|
1.41
|
|
|
|
|
|
|
Stretch-Bend
|
0.1253
|
|
|
|
|
|
|
Torsion
|
2.3169
|
|
|
|
|
|
|
Non-1,4 VDW
|
-0.034
|
|
|
|
|
|
|
1,4 VDW
|
2.8495
|
|
|
|
|
|
|
Total
|
6.8968
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Stretch
|
Length
|
R(0) preferred
|
K(S)
|
Energy
|
|
|
|
C(2)-C(7)
|
1.543
|
1.523
|
4.4
|
0.122
|
central bond
|
|
|
C(1)-C(2)
|
1.534
|
1.523
|
4.4
|
0.040
|
|
|
|
C(7)-C(11)
|
1.534
|
1.523
|
4.4
|
0.040
|
|
|
|
C(2)-H(8)
|
1.117
|
1.113
|
4.6
|
0.007
|
|
|
|
C(7)-H(9)
|
1.117
|
1.113
|
4.6
|
0.007
|
|
|
|
C(2)-H(6)
|
1.117
|
1.113
|
4.6
|
0.007
|
|
|
|
C(7)-H(10)
|
1.117
|
1.113
|
4.6
|
0.007
|
|
|
|
C(1)-H(5)
|
1.115
|
1.113
|
4.6
|
0.001
|
|
|
|
C(11)-H(12)
|
1.115
|
1.113
|
4.6
|
0.001
|
|
|
|
C(1)-H(3)
|
1.113
|
1.113
|
4.6
|
0.000
|
|
|
|
C(1)-H(4)
|
1.113
|
1.113
|
4.6
|
0.000
|
|
|
|
C(11)-H(13)
|
1.113
|
1.113
|
4.6
|
0.000
|
|
|
|
C(11)-H(14)
|
1.113
|
1.113
|
4.6
|
0.000
|
|
|
|
|
|
|
|
|
|
|
|
1,4 van der Waals
|
R
|
RV preferred
|
K(V) 1,4
|
Energy
|
|
|
|
H(8),H(9)
|
2.275
|
3
|
0.047
|
0.692
|
eclipsed Hs
|
These strains have
|
|
H(6),H(10)
|
2.276
|
3
|
0.047
|
0.690
|
been reduced by
|
|
C(1),C(11)
|
2.908
|
3.8
|
0.044
|
0.402
|
eclipsed Cs
|
stretching/bending
|
|
H(9),H(12)
|
2.484
|
3
|
0.047
|
0.227
|
|
|
|
H(5),H(8)
|
2.486
|
3
|
0.047
|
0.224
|
|
|
|
H(5),H(6)
|
2.486
|
3
|
0.047
|
0.224
|
|
|
|
H(10),H(12)
|
2.488
|
3
|
0.047
|
0.222
|
|
|
|
H(10),H(13)
|
2.531
|
3
|
0.047
|
0.166
|
|
|
|
H(4),H(6)
|
2.532
|
3
|
0.047
|
0.164
|
|
|
|
H(3),H(8)
|
2.533
|
3
|
0.047
|
0.164
|
|
|
|
H(9),H(14)
|
2.534
|
3
|
0.047
|
0.162
|
|
|
|
C(2),H(14)
|
2.915
|
3.34
|
0.046
|
0.029
|
|
|
|
C(7),H(3)
|
2.917
|
3.34
|
0.046
|
0.028
|
|
|
|
C(7),H(4)
|
2.918
|
3.34
|
0.046
|
0.028
|
|
|
|
C(2),H(13)
|
2.919
|
3.34
|
0.046
|
0.027
|
|
|
|
H(6),H(9)
|
2.89
|
3
|
0.047
|
-0.034
|
|
|
|
H(8),H(10)
|
2.89
|
3
|
0.047
|
-0.034
|
|
|
|
C(2),H(12)
|
3.566
|
3.34
|
0.046
|
-0.051
|
|
|
|
C(7),H(5)
|
3.566
|
3.34
|
0.046
|
-0.051
|
|
|
|
C(1),H(9)
|
3.358
|
3.34
|
0.046
|
-0.053
|
|
|
|
C(1),H(10)
|
3.359
|
3.34
|
0.046
|
-0.053
|
|
|
|
C(11),H(6)
|
3.358
|
3.34
|
0.046
|
-0.053
|
|
|
|
C(11),H(8)
|
3.359
|
3.34
|
0.046
|
-0.053
|
|
|
|
H(3),H(6)
|
3.104
|
3
|
0.047
|
-0.054
|
|
|
|
H(4),H(8)
|
3.104
|
3
|
0.047
|
-0.054
|
|
|
|
H(9),H(13)
|
3.104
|
3
|
0.047
|
-0.054
|
|
|
|
H(10),H(14)
|
3.105
|
3
|
0.047
|
-0.054
|
|
|
|
|
|
|
|
|
|
|
|
Non 1,4
|
R
|
R preferred
|
K(V) non 1,4
|
Energy
|
|
|
|
H(3),H(14)
|
2.367
|
3
|
0.047
|
0.196
|
Hs on opposing
|
|
|
H(4),H(13)
|
2.376
|
3
|
0.047
|
0.183
|
methyl groups
|
|
|
C(1),H(14)
|
2.848
|
3.34
|
0.046
|
0.052
|
(Also reduced by
|
|
|
C(11),H(3)
|
2.852
|
3.34
|
0.046
|
0.049
|
stretching/bending)
|
|
|
C(11),H(4)
|
2.853
|
3.34
|
0.046
|
0.049
|
|
|
|
C(1),H(13)
|
2.857
|
3.34
|
0.046
|
0.047
|
|
|
|
H(5),H(12)
|
5.126
|
3
|
0.047
|
-0.005
|
|
|
|
H(5),H(9)
|
4.224
|
3
|
0.047
|
-0.016
|
|
|
|
H(5),H(10)
|
4.225
|
3
|
0.047
|
-0.016
|
|
|
|
H(6),H(12)
|
4.225
|
3
|
0.047
|
-0.016
|
|
|
|
H(8),H(12)
|
4.223
|
3
|
0.047
|
-0.016
|
|
|
|
H(5),H(13)
|
3.924
|
3
|
0.047
|
-0.023
|
|
|
|
H(4),H(12)
|
3.921
|
3
|
0.047
|
-0.023
|
|
|
|
H(3),H(12)
|
3.918
|
3
|
0.047
|
-0.023
|
|
|
|
H(5),H(14)
|
3.915
|
3
|
0.047
|
-0.023
|
|
|
|
H(8),H(13)
|
3.88
|
3
|
0.047
|
-0.026
|
|
|
|
H(3),H(10)
|
3.877
|
3
|
0.047
|
-0.026
|
|
|
|
H(4),H(9)
|
3.877
|
3
|
0.047
|
-0.026
|
|
|
|
H(6),H(14)
|
3.875
|
3
|
0.047
|
-0.026
|
|
|
|
C(1),H(12)
|
4.018
|
3.34
|
0.046
|
-0.034
|
|
|
|
C(11),H(5)
|
4.018
|
3.34
|
0.046
|
-0.034
|
|
|
|
H(4),H(10)
|
3.437
|
3
|
0.047
|
-0.042
|
|
|
|
H(6),H(13)
|
3.438
|
3
|
0.047
|
-0.042
|
|
|
|
H(3),H(9)
|
3.436
|
3
|
0.047
|
-0.042
|
|
|
|
H(8),H(14)
|
3.436
|
3
|
0.047
|
-0.042
|
|
|
|
H(4),H(14)
|
2.978
|
3
|
0.047
|
-0.054
|
|
|
|
H(3),H(13)
|
2.988
|
3
|
0.047
|
-0.054
|
|
|
|
|
|
|
|
|
|
|
|
Angle bend
|
Angle
|
preferred
|
K(B)
|
EBend
|
K(St-Bend)
|
ESB
|
|
C(2)-C(7)-C(11)
|
116.408
|
109.5
|
0.45
|
0.471
|
0.12
|
0.0652
|
|
C(1)-C(2)-C(7)
|
116.408
|
109.5
|
0.45
|
0.471
|
0.12
|
0.0652
|
|
H(6)-C(2)-H(8)
|
105.65
|
109.4
|
0.32
|
0.099
|
|
|
|
H(10)-C(7)-H(9)
|
105.646
|
109.4
|
0.32
|
0.099
|
|
|
|
H(12)-C(11)-H(13)
|
106.961
|
109
|
0.32
|
0.029
|
|
|
|
H(4)-C(1)-H(5)
|
106.964
|
109
|
0.32
|
0.029
|
|
|
|
H(3)-C(1)-H(5)
|
106.982
|
109
|
0.32
|
0.029
|
|
|
|
H(12)-C(11)-H(14)
|
106.984
|
109
|
0.32
|
0.029
|
|
|
|
C(7)-C(11)-H(14)
|
111.609
|
110
|
0.36
|
0.020
|
0.09
|
0.0042
|
|
C(2)-C(1)-H(4)
|
111.602
|
110
|
0.36
|
0.020
|
0.09
|
0.0042
|
|
C(2)-C(1)-H(3)
|
111.588
|
110
|
0.36
|
0.020
|
0.09
|
0.0042
|
|
C(7)-C(11)-H(13)
|
111.582
|
110
|
0.36
|
0.020
|
0.09
|
0.0042
|
|
C(11)-C(7)-H(9)
|
107.977
|
109.41
|
0.36
|
0.016
|
0.09
|
-0.0051
|
|
C(1)-C(2)-H(6)
|
107.984
|
109.41
|
0.36
|
0.016
|
0.09
|
-0.0051
|
|
C(1)-C(2)-H(8)
|
107.998
|
109.41
|
0.36
|
0.016
|
0.09
|
-0.005
|
|
C(11)-C(7)-H(10)
|
108.003
|
109.41
|
0.36
|
0.016
|
0.09
|
-0.005
|
|
C(2)-C(1)-H(5)
|
110.755
|
110
|
0.36
|
0.005
|
0.09
|
0.0022
|
|
C(7)-C(11)-H(12)
|
110.754
|
110
|
0.36
|
0.005
|
0.09
|
0.0022
|
|
C(2)-C(7)-H(9)
|
109.117
|
109.41
|
0.36
|
0.001
|
0.09
|
-0.0016
|
|
C(7)-C(2)-H(6)
|
109.129
|
109.41
|
0.36
|
0.001
|
0.09
|
-0.0016
|
|
C(7)-C(2)-H(8)
|
109.143
|
109.41
|
0.36
|
0.001
|
0.09
|
-0.0015
|
|
H(3)-C(1)-H(4)
|
108.732
|
109
|
0.32
|
0.001
|
|
|
|
C(2)-C(7)-H(10)
|
109.157
|
109.41
|
0.36
|
0.001
|
0.09
|
-0.0014
|
|
H(13)-C(11)-H(14)
|
108.732
|
109
|
0.32
|
0.001
|
|
|
|
|
|
|
|
|
|
|
|
Torsions
|
angle
|
V1
|
V2
|
V3
|
E torsion
|
|
|
C(1)-C(2)-C(7)-C(11)
|
0
|
0.2
|
0.27
|
0.093
|
0.293
|
All these
|
|
C(1)-C(2)-C(7)-H(9)
|
-122.469
|
0
|
0
|
0.267
|
0.266
|
values
|
|
H(6)-C(2)-C(7)-C(11)
|
-122.484
|
0
|
0
|
0.267
|
0.266
|
involve
|
|
C(1)-C(2)-C(7)-H(10)
|
122.535
|
0
|
0
|
0.267
|
0.266
|
torsion
|
|
H(8)-C(2)-C(7)-C(11)
|
122.518
|
0
|
0
|
0.267
|
0.266
|
about the
|
|
H(6)-C(2)-C(7)-H(10)
|
0
|
0
|
0
|
0.237
|
0.237
|
central (C2-C7)
|
|
H(8)-C(2)-C(7)-H(9)
|
0
|
0
|
0
|
0.237
|
0.237
|
bond.
|
|
H(6)-C(2)-C(7)-H(9)
|
115.05
|
0
|
0
|
0.237
|
0.233
|
They sum to
|
|
H(8)-C(2)-C(7)-H(10)
|
-114.95
|
0
|
0
|
0.237
|
0.233
|
2.3 kcal
|
|
H(9)-C(7)-C(11)-H(13)
|
-175.683
|
0
|
0
|
0.237
|
0.003
|
|
|
H(4)-C(1)-C(2)-H(8)
|
175.923
|
0
|
0
|
0.237
|
0.003
|
|
|
H(3)-C(1)-C(2)-H(6)
|
-176.009
|
0
|
0
|
0.237
|
0.003
|
|
|
H(10)-C(7)-C(11)-H(14)
|
176.241
|
0
|
0
|
0.237
|
0.002
|
|
|
H(9)-C(7)-C(11)-H(12)
|
-56.638
|
0
|
0
|
0.237
|
0.002
|
|
|
H(5)-C(1)-C(2)-H(8)
|
56.864
|
0
|
0
|
0.237
|
0.002
|
|
|
H(5)-C(1)-C(2)-H(6)
|
-56.941
|
0
|
0
|
0.237
|
0.002
|
|
|
H(10)-C(7)-C(11)-H(12)
|
57.165
|
0
|
0
|
0.237
|
0.001
|
|
|
H(9)-C(7)-C(11)-H(14)
|
62.442
|
0
|
0
|
0.237
|
0.001
|
|
|
H(3)-C(1)-C(2)-H(8)
|
-62.2
|
0
|
0
|
0.237
|
0.001
|
|
|
H(4)-C(1)-C(2)-H(6)
|
62.115
|
0
|
0
|
0.237
|
0.001
|
|
|
H(10)-C(7)-C(11)-H(13)
|
-61.873
|
0
|
0
|
0.237
|
0.001
|
|
|
C(2)-C(7)-C(11)-H(13)
|
61.26
|
0
|
0
|
0.267
|
0.000
|
|
|
H(3)-C(1)-C(2)-C(7)
|
60.913
|
0
|
0
|
0.267
|
0.000
|
|
|
H(4)-C(1)-C(2)-C(7)
|
-60.966
|
0
|
0
|
0.267
|
0.000
|
|
|
C(2)-C(7)-C(11)-H(14)
|
-60.62
|
0
|
0
|
0.267
|
0.000
|
|
|
H(5)-C(1)-C(2)-C(7)
|
180
|
0
|
0
|
0.267
|
0.000
|
|
|
C(2)-C(7)-C(11)-H(12)
|
-179.684
|
0
|
0
|
0.267
|
0.000
|
|
If the concept of
storing more energy in the easier distortion seems funny to you,
think of hooking a rubber band to a suspension spring of a Mack
truck and pulling on the end. The rubber band would stretch and
the spring wouldn't, so all the energy would go into the rubber
band.
If you want to be more concrete and
quantitative, think of it this way:
The force on a spring depends on how much it is
stretched (dx). The energy stored in the spring depends on the
square of the distortion (this is why the potential for an
harmonic oscillator is a parabola). Where k is the spring
constant,
F = -k dx ; E = 1/2 k
(dx)2
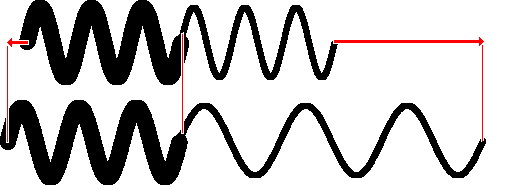
When two springs interact, as when you hook a
weak spring ( k2) to a strong spring ( k1 =
8 x
k2) and pull on the two ends, the force on the two
springs is the same, so the weak one stretches more (here 8 times
as much):
F2 = (-)F1 means
that k2 dx2 = k1 dx1
means that dx2 = 8 dx1
This means that the weaker spring stores 8
times as much energy as the stronger spring (k is 8 times smaller,
but dx2 is 64 times larger).
In summary: since the forces must
balance, more energy goes into the weaker
spring.
copyright 2001 J. M. McBride